#include <species.hpp>
Public Types | |
| enum class | LaunchBounds { Default , Custom } |
Public Member Functions | |
| Species (int idx_in, int nonadiabatic_idx_in, bool is_electron_in, bool is_adiabatic_in, KinType kintype_in, double mass_in, double charge_in, double charge_eu_in, int ncycles_in) | |
| Species (NLReader::NamelistReader &nlr, const Grid< DeviceType > &grid, const MagneticField< DeviceType > &magnetic_field, const DomainDecomposition< DeviceType > &pol_decomp, int idx_in, int nonadiabatic_idx_in) | |
| Species (SpeciesType sp_type, int n_ptl) | |
| Species (int n_ptl_in) | |
| void | resize_particles (int new_n_ptl) |
| void | resize_host_particles_to_match_device () |
| void | unassign_host_particles () |
| void | resize_device_particles () |
| void | resize_device_particles (int new_n_ptl) |
| void | copy_particles_to_device () |
| void | copy_particles_from_device () |
| void | copy_particles_to_device_if_resident () |
| void | copy_particles_from_device_if_resident () |
| void | copy_particles_to_device_if_not_resident () |
| void | copy_particles_from_device_if_not_resident () |
| void | set_buffer_particles_d () |
| void | set_buffer_phase0_d () |
| template<typename F > | |
| void | for_all_particles (const std::string label, F lambda_func) const |
| void | back_up_SoA (Cabana::AoSoA< ParticleDataTypes, Device, VEC_LEN > &backup_SoA, int offset, int n) const |
| void | restore_backup_SoA (Cabana::AoSoA< ParticleDataTypes, Device, VEC_LEN > &backup_SoA, int offset, int n) const |
| template<typename F > | |
| void | for_particle_range (int begin_idx, int end_idx, const std::string label, F lambda_func) const |
| template<typename F > | |
| void | for_all_particles (const std::string label, F lambda_func, const PtlMvmt mvmt, LaunchBounds launch_bounds=LaunchBounds::Default) |
| KOKKOS_INLINE_FUNCTION VecParticles * | ptl () const |
| KOKKOS_INLINE_FUNCTION VecPhase * | ph0 () const |
| void | copy_to_phase0 (Species< Device > &species) |
| bool | phase0_is_stored () const |
| void | copy_phase0_to_device_if_not_resident () |
| void | save_backup_particles () |
| void | restore_particles_from_backup () |
| void | move_phase0_from_device_if_not_resident () |
| void | clear_backup_phase () |
| KOKKOS_INLINE_FUNCTION void | restore_phase_from_phase0 (const AoSoAIndices< Device > &inds, SimdParticles &part_one) const |
| long long int | get_total_n_ptl () |
| int | get_max_n_ptl () |
| KOKKOS_INLINE_FUNCTION double | get_fg_gyro_radius (int inode, double smu_n, double bfield) const |
| KOKKOS_INLINE_FUNCTION double | get_f0_eq_thermal_velocity (int inode) const |
| KOKKOS_INLINE_FUNCTION double | get_f0_eq_thermal_velocity_lnode (int inode) const |
| KOKKOS_INLINE_FUNCTION double | get_f0_eq_thermal_velocity_lnode_h (int inode) const |
| KOKKOS_INLINE_FUNCTION double | get_f0_fg_unit_velocity_lnode_h (int inode) const |
| KOKKOS_INLINE_FUNCTION void | get_particle_velocity_and_nearest_node (const Grid< DeviceType > &grid, const MagneticField< DeviceType > &magnetic_field, const DomainDecomposition< DeviceType > &pol_decomp, SimdParticles &part, Simd< double > &smu, Simd< double > &vp, Simd< int > &nearest_node, Simd< bool > ¬_in_triangle, Simd< bool > ¬_in_poloidal_domain) const |
| KOKKOS_INLINE_FUNCTION double | eq_flow_ms (const MagneticField< DeviceType > &magnetic_field, double psi_in, double r, double z, double bphi_over_b) const |
| KOKKOS_INLINE_FUNCTION void | get_tr_save (int i_item, Simd< int > &itr) const |
| void | update_decomposed_f0_calculations (const DomainDecomposition< DeviceType > &pol_decomp, const Grid< DeviceType > &grid, const MagneticField< DeviceType > &magnetic_field, const VelocityGrid &vgrid) |
| void | initialize_global_f0_arrays (const Grid< DeviceType > &grid, const MagneticField< DeviceType > &magnetic_field) |
| void | write_ptl_checkpoint_files (const DomainDecomposition< DeviceType > &pol_decomp, const XGC_IO_Stream &stream, std::string sp_name) |
| void | write_f0_checkpoint_files (const DomainDecomposition< DeviceType > &pol_decomp, const XGC_IO_Stream &stream, std::string sp_name) |
| void | read_f0_checkpoint_files (const DomainDecomposition< DeviceType > &pol_decomp, const XGC_IO_Stream &stream, std::string sp_name) |
| void | read_ptl_checkpoint_files (const DomainDecomposition< DeviceType > &pol_decomp, const XGC_IO_Stream &stream, std::string sp_name, bool n_ranks_is_same, int version) |
| void | read_initial_distribution (NLReader::NamelistReader &nlr, const DomainDecomposition< DeviceType > &pol_decomp) |
| long long int | get_max_gid () const |
| void | get_ptl_write_total_and_offsets (const DomainDecomposition< DeviceType > &pol_decomp, long long int &inum_total, long long int &ioff) const |
Static Public Member Functions | |
| static std::vector< MemoryPrediction > | estimate_memory_usage (NLReader::NamelistReader &nlr, const Grid< DeviceType > &grid, const DomainDecomposition< DeviceType > &pol_decomp, int species_idx) |
| static int | get_initial_n_ptl (NLReader::NamelistReader &nlr, const Grid< DeviceType > &grid, const DomainDecomposition< DeviceType > &pol_decomp, int species_idx, bool verbose) |
Public Attributes | |
| int | idx |
| Index in all_species. More... | |
| bool | is_electron |
| Whether this species is the electrons. More... | |
| bool | is_adiabatic |
| Whether this species is adiabatic. More... | |
| int | nonadiabatic_idx |
| Index of species skipping adiabatic species (for compatibility with fortran arrays) More... | |
| KinType | kintype |
| Whether the species is gyrokinetic or drift kinetic. More... | |
| double | mass |
| Particle mass. More... | |
| double | charge |
| Particle charge. More... | |
| double | charge_eu |
| Particle charge in eu. More... | |
| double | c_m |
| c/m More... | |
| double | c2_2m |
| c2/2m More... | |
| MarkerType | marker_type |
| Marker type: reduced delta-f, total-f, full-f, or none (placeholder for adiabatic species) More... | |
| FAnalyticShape | f_analytic_shape |
| f_analytic_shape shape: Maxwellian, SlowingDown or None More... | |
| WeightEvoEq | weight_evo_eq |
| bool | dynamic_f0 |
| Whether f0 can evolve in time. More... | |
| bool | maxwellian_init |
| whether initial distribution is maxwellian More... | |
| int | ncycles |
| Number of subcycles. More... | |
| int | ncycles_between_sorts |
| Number of subcycles between sorts. More... | |
| int | minimum_ptl_reservation |
| The minimum reservation size for particles. More... | |
| int | n_ptl |
| Number of particles. More... | |
| Cabana::AoSoA< ParticleDataTypes, HostType, VEC_LEN > | particles |
| Particles. More... | |
| Cabana::AoSoA< ParticleDataTypes, Device, VEC_LEN > | particles_d |
| Particles on device. More... | |
| bool | owns_particles_d |
| Whether the species owns the device particle allocation right now. More... | |
| bool | particles_resident_on_device |
| Whether the particles can reside on device. More... | |
| bool | stream_particles |
| Whether to stream particles between host and device if possible. More... | |
| RKRestorationMethod | RK_restoration_method |
| Currently, electrons must use first method and ions must use second. More... | |
| bool | particles_are_backed_up |
| Whether particles are currently backed up. More... | |
| Cabana::AoSoA< PhaseDataTypes, HostType, VEC_LEN > | phase0 |
| Cabana::AoSoA< PhaseDataTypes, Device, VEC_LEN > | phase0_d |
| Cabana::AoSoA< ParticleDataTypes, HostType, VEC_LEN > | backup_particles |
| Copy of particles to be restored for RK2. More... | |
| Cabana::AoSoA< ParticleDataTypes, DeviceType, VEC_LEN > | backup_particles_d |
| Copy of particles to be restored for RK2. More... | |
| int | n_backup_particles |
| bool | backup_particles_on_device |
| View< int *, CLayout, Device > | tr_save |
| Distribution< Device > | f0 |
| Species distribution in velocity space on local mesh nodes. More... | |
| int | collision_grid_index |
| Which collision grid to use. More... | |
| Eq::Profile< Device > | eq_temp |
| Eq::Profile< Device > | eq_den |
| Eq::Profile< Device > | eq_flow |
| int | eq_flow_type |
| Eq::Profile< Device > | eq_fg_temp |
| Eq::Profile< Device > | eq_fg_flow |
| int | eq_fg_flow_type |
| Eq::Profile< Device > | eq_mk_temp |
| Eq::Profile< Device > | eq_mk_den |
| Eq::Profile< Device > | eq_mk_flow |
| int | eq_mk_flow_type |
| GyroAverageMatrices< Device > | gyro_avg_matrices |
Member Enumeration Documentation
◆ LaunchBounds
|
strong |
Constructor & Destructor Documentation
◆ Species() [1/4]
| Species< Device >::Species | ( | int | idx_in, |
| int | nonadiabatic_idx_in, | ||
| bool | is_electron_in, | ||
| bool | is_adiabatic_in, | ||
| KinType | kintype_in, | ||
| double | mass_in, | ||
| double | charge_in, | ||
| double | charge_eu_in, | ||
| int | ncycles_in | ||
| ) |
Constructor for species class used in collisions kernel
◆ Species() [2/4]
| Species< Device >::Species | ( | NLReader::NamelistReader & | nlr, |
| const Grid< DeviceType > & | grid, | ||
| const MagneticField< DeviceType > & | magnetic_field, | ||
| const DomainDecomposition< DeviceType > & | pol_decomp, | ||
| int | idx_in, | ||
| int | nonadiabatic_idx_in | ||
| ) |
Constructor for species class
< Specifies the weight evolution equation method: Direct, PDE, or None.
< - Direct integration: Is used in the Total-F method, where the source term and particle contributions are evaluated separately. Refer to Section II C of Hager et al., Phys. Plasmas 29, 112308 (2022) for more details.
- PDE: The PDE (Partial Differential Equation) method, where the source term and particle contributions are evaluated together (delta-f).
- None: No weight evolution is applied.
◆ Species() [3/4]
|
inline |
◆ Species() [4/4]

Member Function Documentation
◆ back_up_SoA()
|
inline |

◆ clear_backup_phase()
|
inline |

◆ copy_particles_from_device()
|
inline |

◆ copy_particles_from_device_if_not_resident()
|
inline |

◆ copy_particles_from_device_if_resident()
|
inline |

◆ copy_particles_to_device()
|
inline |
◆ copy_particles_to_device_if_not_resident()
|
inline |
◆ copy_particles_to_device_if_resident()
|
inline |
◆ copy_phase0_to_device_if_not_resident()
|
inline |
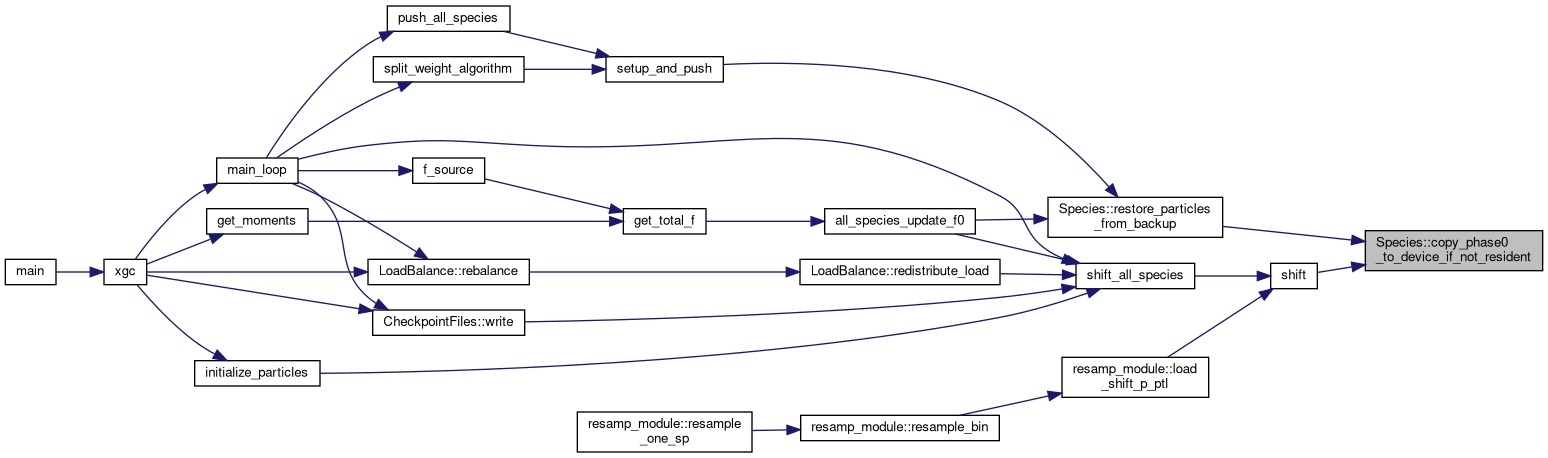
◆ copy_to_phase0()
|
inline |
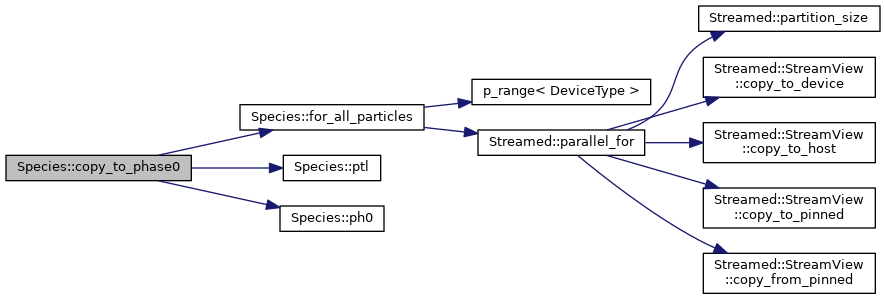

◆ eq_flow_ms()
|
inline |
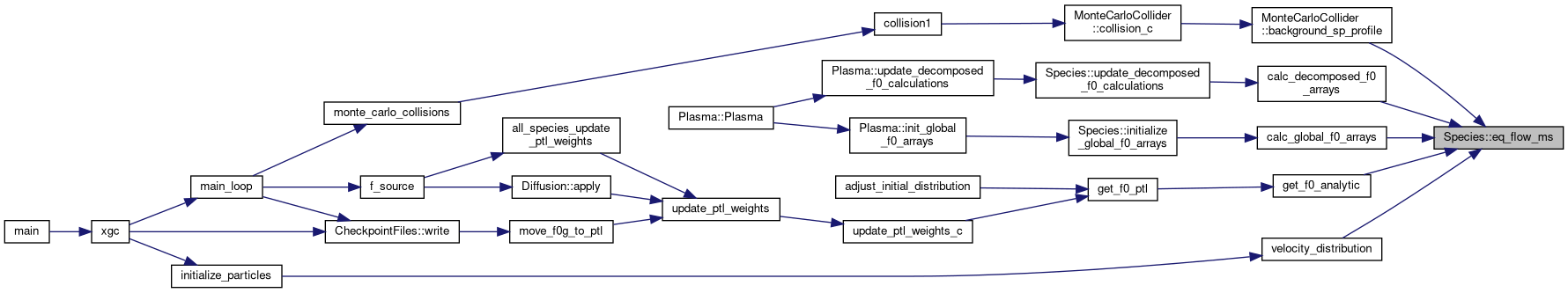
◆ estimate_memory_usage()
|
static |
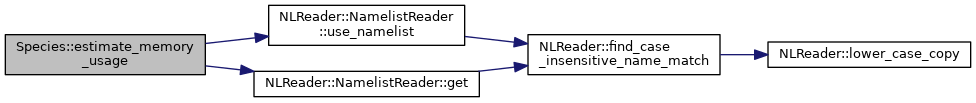

◆ for_all_particles() [1/2]
|
inline |
Loops over particles and calls a parallel_for, with no particle movement
- Parameters
-
[in] label is the label of the parallel_for used for debugging [in] lambda_func is the lambda function. Must be copy [=] not by reference
◆ for_all_particles() [2/2]
|
inline |
Loops over particles and calls a parallel_for, with particle being moved where requested
- Parameters
-
[in] label is the label of the parallel_for used for debugging [in] lambda_func is the lambda function. Must be copy [=] not by reference [in] mvmt specifies whether particles need to be sent to device or host [in] launch_bounds specifies whether to use default launch bounds or custom
◆ for_particle_range()
|
inline |
Loops over a subset of particles and calls a parallel_for, with no particle movement
- Parameters
-
[in] label is the label of the parallel_for used for debugging [in] lambda_func is the lambda function. Must be copy [=] not by reference
◆ get_f0_eq_thermal_velocity()
|
inline |

◆ get_f0_eq_thermal_velocity_lnode()
|
inline |
◆ get_f0_eq_thermal_velocity_lnode_h()
|
inline |
◆ get_f0_fg_unit_velocity_lnode_h()
|
inline |
◆ get_fg_gyro_radius()
|
inline |
◆ get_initial_n_ptl()
|
static |
◆ get_max_gid()
| long long int Species< Device >::get_max_gid |

◆ get_max_n_ptl()
|
inline |

◆ get_particle_velocity_and_nearest_node()
|
inline |
◆ get_ptl_write_total_and_offsets()
| void Species< Device >::get_ptl_write_total_and_offsets | ( | const DomainDecomposition< DeviceType > & | pol_decomp, |
| long long int & | inum_total, | ||
| long long int & | ioff | ||
| ) | const |
◆ get_total_n_ptl()
|
inline |
◆ get_tr_save()
|
inline |
◆ initialize_global_f0_arrays()
| void Species< Device >::initialize_global_f0_arrays | ( | const Grid< DeviceType > & | grid, |
| const MagneticField< DeviceType > & | magnetic_field | ||
| ) |
◆ move_phase0_from_device_if_not_resident()
|
inline |
◆ ph0()
◆ phase0_is_stored()
|
inline |
◆ ptl()
|
inline |
◆ read_f0_checkpoint_files()
| void Species< Device >::read_f0_checkpoint_files | ( | const DomainDecomposition< DeviceType > & | pol_decomp, |
| const XGC_IO_Stream & | stream, | ||
| std::string | sp_name | ||
| ) |


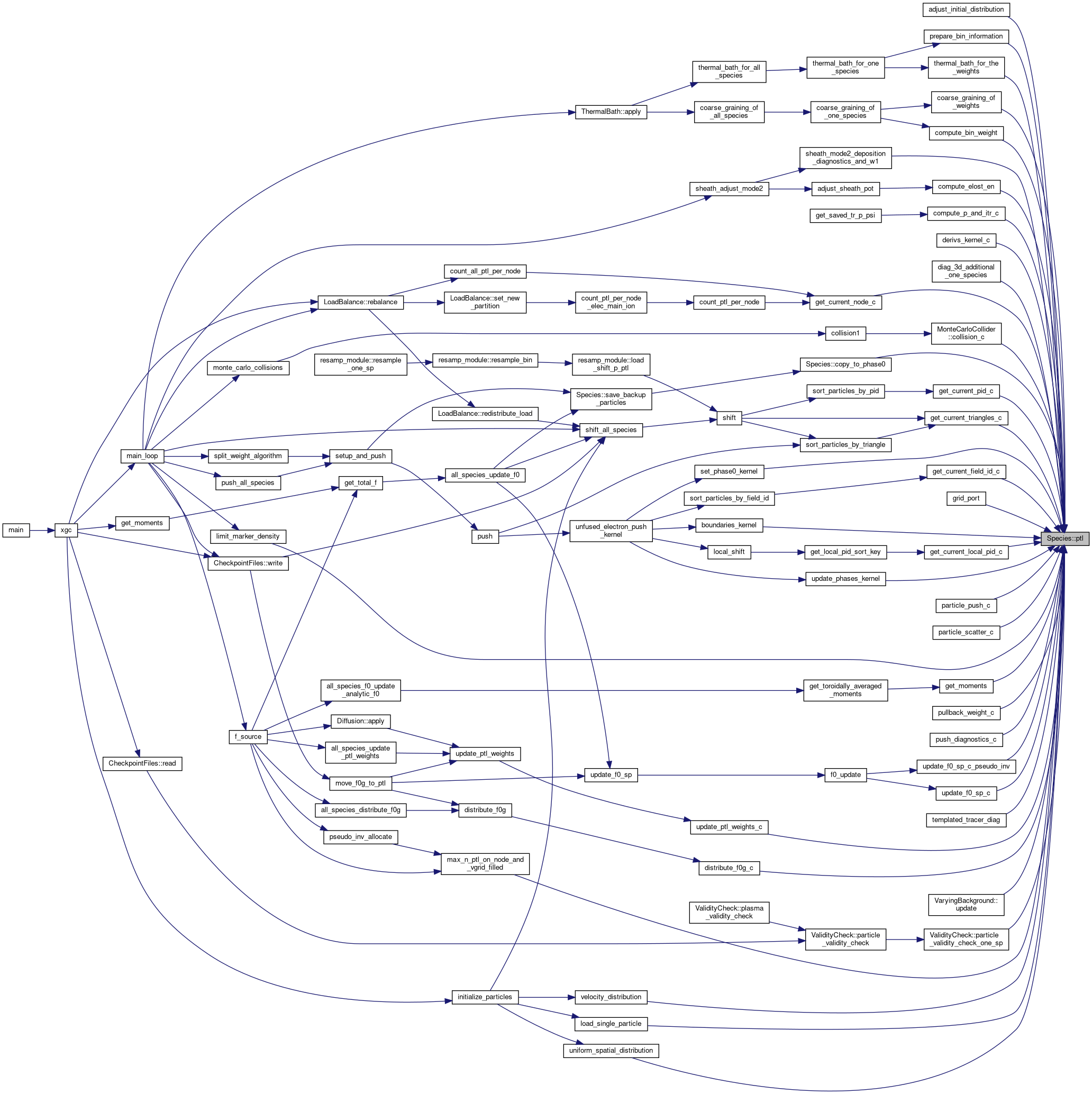
◆ read_initial_distribution()
| void Species< Device >::read_initial_distribution | ( | NLReader::NamelistReader & | nlr, |
| const DomainDecomposition< DeviceType > & | pol_decomp | ||
| ) |
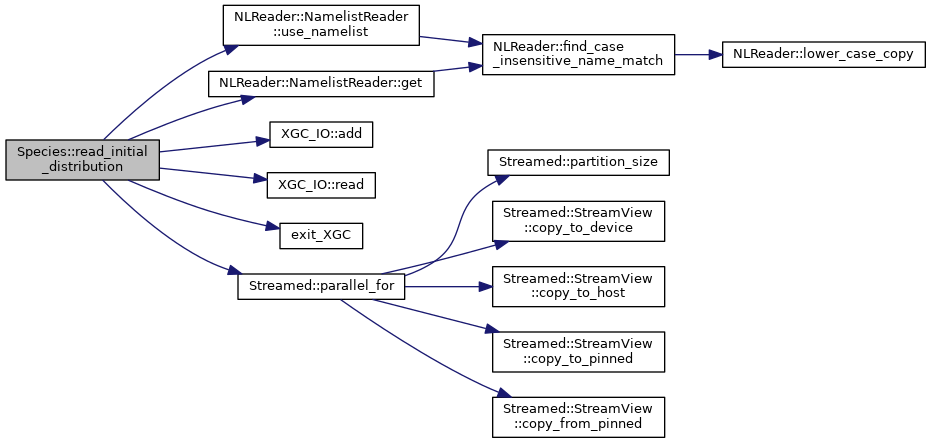
◆ read_ptl_checkpoint_files()
| void Species< Device >::read_ptl_checkpoint_files | ( | const DomainDecomposition< DeviceType > & | pol_decomp, |
| const XGC_IO_Stream & | stream, | ||
| std::string | sp_name, | ||
| bool | n_ranks_is_same, | ||
| int | version | ||
| ) |

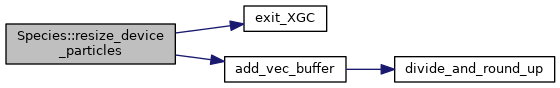
◆ resize_device_particles() [1/2]
|
inline |
◆ resize_device_particles() [2/2]
|
inline |
◆ resize_host_particles_to_match_device()
|
inline |
◆ resize_particles()
|
inline |

◆ restore_backup_SoA()
|
inline |

◆ restore_particles_from_backup()
|
inline |

◆ restore_phase_from_phase0()
|
inline |
◆ save_backup_particles()
|
inline |

◆ set_buffer_particles_d()
|
inline |
Fills the remainder of the last AoSoA vector with realistic-looking particles (clones of the final particle). Since gid is set to -1, they will basically act as tracers in the kernel. Ideally garbage data in this buffer doesn't affect anything, but better to be safe here
- Returns
- void
◆ set_buffer_phase0_d()
|
inline |
Fills the remainder of the last AoSoA phase0 vector with realistic-looking particles (clones of the final particle). Since gid is set to -1, they will basically act as tracers in the kernel. Ideally garbage data in this buffer doesn't affect anything, but better to be safe here
- Returns
- void
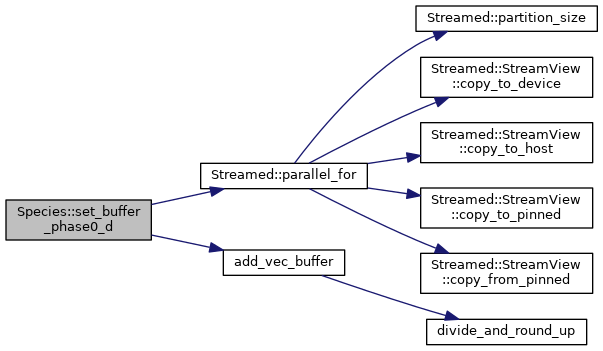

◆ unassign_host_particles()
|
inline |
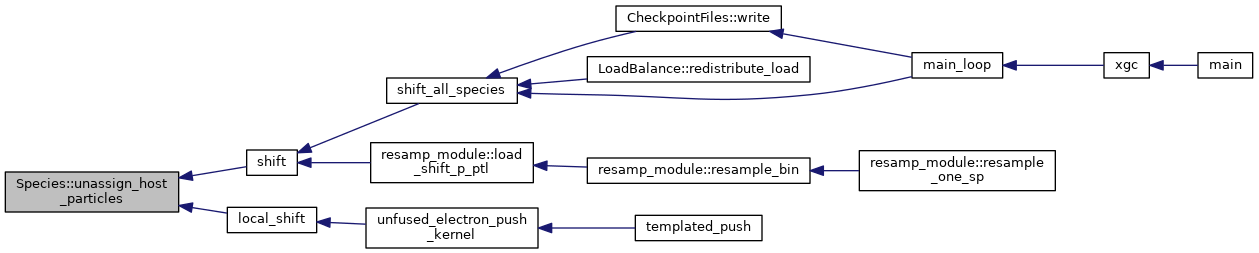
◆ update_decomposed_f0_calculations()
| void Species< Device >::update_decomposed_f0_calculations | ( | const DomainDecomposition< DeviceType > & | pol_decomp, |
| const Grid< DeviceType > & | grid, | ||
| const MagneticField< DeviceType > & | magnetic_field, | ||
| const VelocityGrid & | vgrid | ||
| ) |
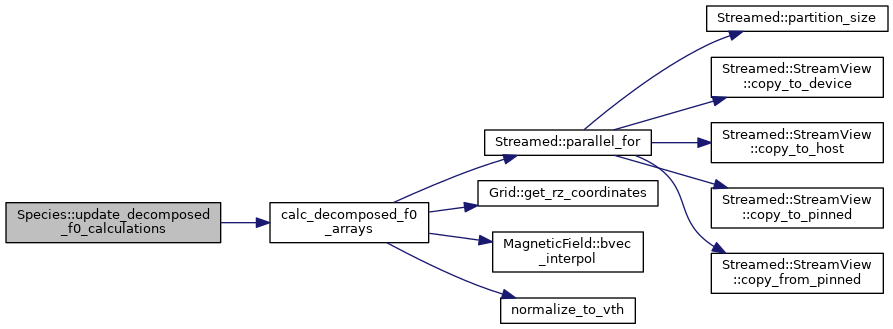
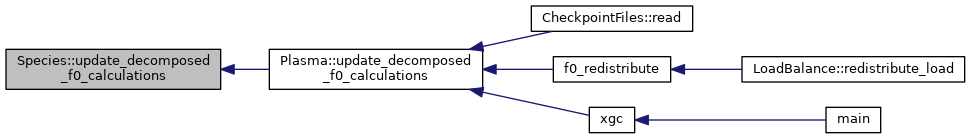
◆ write_f0_checkpoint_files()
| void Species< Device >::write_f0_checkpoint_files | ( | const DomainDecomposition< DeviceType > & | pol_decomp, |
| const XGC_IO_Stream & | stream, | ||
| std::string | sp_name | ||
| ) |


◆ write_ptl_checkpoint_files()
| void Species< Device >::write_ptl_checkpoint_files | ( | const DomainDecomposition< DeviceType > & | pol_decomp, |
| const XGC_IO_Stream & | stream, | ||
| std::string | sp_name | ||
| ) |

Member Data Documentation
◆ backup_particles
| Cabana::AoSoA<ParticleDataTypes,HostType,VEC_LEN> Species< Device >::backup_particles |
Copy of particles to be restored for RK2.
◆ backup_particles_d
| Cabana::AoSoA<ParticleDataTypes,DeviceType,VEC_LEN> Species< Device >::backup_particles_d |
Copy of particles to be restored for RK2.
◆ backup_particles_on_device
| bool Species< Device >::backup_particles_on_device |
◆ c2_2m
| double Species< Device >::c2_2m |
c2/2m
◆ c_m
| double Species< Device >::c_m |
c/m
◆ charge
| double Species< Device >::charge |
Particle charge.
◆ charge_eu
| double Species< Device >::charge_eu |
Particle charge in eu.
◆ collision_grid_index
| int Species< Device >::collision_grid_index |
Which collision grid to use.
◆ dynamic_f0
| bool Species< Device >::dynamic_f0 |
Whether f0 can evolve in time.
◆ eq_den
| Eq::Profile<Device> Species< Device >::eq_den |
◆ eq_fg_flow
| Eq::Profile<Device> Species< Device >::eq_fg_flow |
◆ eq_fg_flow_type
| int Species< Device >::eq_fg_flow_type |
◆ eq_fg_temp
| Eq::Profile<Device> Species< Device >::eq_fg_temp |
◆ eq_flow
| Eq::Profile<Device> Species< Device >::eq_flow |
◆ eq_flow_type
| int Species< Device >::eq_flow_type |
◆ eq_mk_den
| Eq::Profile<Device> Species< Device >::eq_mk_den |
◆ eq_mk_flow
| Eq::Profile<Device> Species< Device >::eq_mk_flow |
◆ eq_mk_flow_type
| int Species< Device >::eq_mk_flow_type |
◆ eq_mk_temp
| Eq::Profile<Device> Species< Device >::eq_mk_temp |
◆ eq_temp
| Eq::Profile<Device> Species< Device >::eq_temp |
◆ f0
| Distribution<Device> Species< Device >::f0 |
Species distribution in velocity space on local mesh nodes.
◆ f_analytic_shape
| FAnalyticShape Species< Device >::f_analytic_shape |
f_analytic_shape shape: Maxwellian, SlowingDown or None
◆ gyro_avg_matrices
| GyroAverageMatrices<Device> Species< Device >::gyro_avg_matrices |
◆ idx
| int Species< Device >::idx |
Index in all_species.
◆ is_adiabatic
| bool Species< Device >::is_adiabatic |
Whether this species is adiabatic.
◆ is_electron
| bool Species< Device >::is_electron |
Whether this species is the electrons.
◆ kintype
Whether the species is gyrokinetic or drift kinetic.
◆ marker_type
| MarkerType Species< Device >::marker_type |
Marker type: reduced delta-f, total-f, full-f, or none (placeholder for adiabatic species)
◆ mass
| double Species< Device >::mass |
Particle mass.
◆ maxwellian_init
| bool Species< Device >::maxwellian_init |
whether initial distribution is maxwellian
◆ minimum_ptl_reservation
| int Species< Device >::minimum_ptl_reservation |
The minimum reservation size for particles.
◆ n_backup_particles
| int Species< Device >::n_backup_particles |
◆ n_ptl
| int Species< Device >::n_ptl |
Number of particles.
◆ ncycles
| int Species< Device >::ncycles |
Number of subcycles.
◆ ncycles_between_sorts
| int Species< Device >::ncycles_between_sorts |
Number of subcycles between sorts.
◆ nonadiabatic_idx
| int Species< Device >::nonadiabatic_idx |
Index of species skipping adiabatic species (for compatibility with fortran arrays)
◆ owns_particles_d
| bool Species< Device >::owns_particles_d |
Whether the species owns the device particle allocation right now.
◆ particles
| Cabana::AoSoA<ParticleDataTypes,HostType,VEC_LEN> Species< Device >::particles |
Particles.
◆ particles_are_backed_up
| bool Species< Device >::particles_are_backed_up |
Whether particles are currently backed up.
◆ particles_d
| Cabana::AoSoA<ParticleDataTypes,Device,VEC_LEN> Species< Device >::particles_d |
Particles on device.
◆ particles_resident_on_device
| bool Species< Device >::particles_resident_on_device |
Whether the particles can reside on device.
◆ phase0
| Cabana::AoSoA<PhaseDataTypes,HostType,VEC_LEN> Species< Device >::phase0 |
◆ phase0_d
| Cabana::AoSoA<PhaseDataTypes,Device,VEC_LEN> Species< Device >::phase0_d |
◆ RK_restoration_method
| RKRestorationMethod Species< Device >::RK_restoration_method |
Currently, electrons must use first method and ions must use second.
Whether to restore particles on original rank, or to carry phase0 to new rank and restore there.
◆ stream_particles
| bool Species< Device >::stream_particles |
Whether to stream particles between host and device if possible.
◆ tr_save
◆ weight_evo_eq
| WeightEvoEq Species< Device >::weight_evo_eq |
Specifies the weight evolution equation method: Direct, PDE, or None.
- Direct integration: Is used in the Total-F method, where the source term and particle contributions are evaluated separately. Refer to Section II C of Hager et al., Phys. Plasmas 29, 112308 (2022) for more details.
- PDE: The PDE (Partial Differential Equation) method, where the source term and particle contributions are evaluated together (delta-f).
- None: No weight evolution is applied.
The documentation for this class was generated from the following files:
- /p/test_ssd/builds/t3_84szKM/0/xgc/XGC-Devel/XGC_core/cpp/species.hpp
- /p/test_ssd/builds/t3_84szKM/0/xgc/XGC-Devel/XGC_core/cpp/species.cpp