|
XGCa
|
|
XGCa
|
Public Member Functions | |
| subroutine | init_ksp_comm (nnode) |
| Initializes an MPI communicator for use with PETSc KSP solves (Poisson, Ampere). The size of the KSP comm group is set such that, if possible, the number of equations per MPI rank is larger than 5000, which is roughly the weak scaling rollover. More... | |
| subroutine | petsc_get_sizes (grid, bc, n_equation, n_boundary, xgc_petsc, petsc_xgc_bd) |
| Calculate (i) the number of equations (vertices) of the solver, (ii) the number of XGC boundary vertices included in the solver, (iii) (preliminary) mapping from XGC vertices to PETSc equations, (iv) mapping from XGC vertices to PETSc boundary conditions, all based the XGC boundary object passed as input. More... | |
| subroutine | petsc_get_bd_map (grid, bc, n_boundary, petsc_xgc_bd, petsc_bd_xgc) |
| Generate mapping from PETSc boundary conditions to XGC vertices. This requires the results of petsc_get_sizes because the number of boundary conditions is not known a priori. More... | |
| subroutine | getnnz (grid, nloc, low, high, d_nnz, o_nnz, xgc_petsc, nglobal, ierr) |
| Computes the number of non-zero entries nnz per row in the rank-local rows. More... | |
| subroutine | petsc_get_partitioning (grid, bc, solver_data, comm, num_pe, n_eq_tot_in, set_diffusion_matrix, xgc_petsc, petsc_xgc_bd, n_eq_loc, xgc_proc_out, proc_eq, ierr) |
| Generates a domain partitioning of part of the XGC mesh used by a solver (as defined by the boundary conditions bc) using Parmetis and generates a mapping between PETSc equation index and XGC MPI ranks (those in the communicator comm used by the solver). More... | |
| subroutine | petsc_get_mapping (nnode, bc, comm, num_pe, n_eq_tot, n_eq_loc, xgc_proc, proc_eq, xgc_petsc, petsc_xgc, petscloc_xgc) |
| Based on the Parmetis partition of the XGC domain, updates the mappings between XGC vertices and PETSc equations. More... | |
| subroutine | petsc_get_template_mat (grid, comm, n_eq_tot, n_eq_loc, xgc_petsc, solver_template_mat, ierr) |
| Uses pre-computed (petsc_get_partitioning) local matrix sizes and XGC vertex to PETSc equation mapping to set up a blank template matrix with sufficient pre-allocated memory. More... | |
| subroutine | petsc_get_bc_mat (solver, n_eq_tot, n_eq_loc, ierr) |
| Creates the (empty) LHS and RHS matrices for Dirichlet boundary conditions. More... | |
| subroutine | petsc_get_fsa_mat (solver, n_eq_tot, n_eq_loc, ierr) |
| creates and allocates interior/surface FSA matrices More... | |
| subroutine | petsc_get_fsa_bc_mat (solver, ierr) |
| creates and allocates boundary/surface FSA matrices More... | |
| subroutine | petsc_get_scatter_one_block (solver, nnode, n_eq_tot, n_eq_loc, petsc_xgc, ierr) |
| Generate PETSc scatter mapping from XGC vertices to PETSc equation for a simple solver with one equation (block). Also sets up the corresponding LHS and RHS PETSc vectors. More... | |
| subroutine | petsc_get_scatter_multi_block (nnode, ksp, mat_all_blocks, mat_one_block, blocksize, varnames, n_eq_tot, n_eq_loc, petscloc_xgc, petsc_xgc, iss, to_petsc, from_petsc, ierr) |
| Sets up a PETSc scatter object for a (2D) multi-block field-split solver. More... | |
| subroutine | petsc_to_xgc (nnode, blocksize, from_petsc, field_petsc, field_xgc, ierr) |
| This routine scatters values from the distributed vector compatible with the block-matrix of the 2D diffusion model to local variables on the XGC solver grid. More... | |
| subroutine | petsc_to_xgc_bd (nn, this, x, xvec) |
| Scatters a PETSc vector to XGC boundary data. More... | |
| subroutine | xgc_to_petsc (nnode, blocksize, to_petsc, field_petsc, field_xgc, ierr) |
| This routine scatters values from local variable on the XGC solver grid to the distributed vector compatible with the block-matrix of the 2D diffusion model. More... | |
| subroutine | xgc_to_petsc_bd (nn, this, x, xvec) |
| Scatters XGC boundary data to a PETSc vector. More... | |
| subroutine | create_1field_solver (solver, mat, ierr) |
| Creates a PETSc KSP solver for a one-block system, i.e., for a single equation on the XGC mesh. More... | |
| subroutine | create_axisym_iter_solver (solver, ierr) |
| Creates and sets up the PETSc KSP solver and preconditioner for inverting the axisymmetric Poisson operator. More... | |
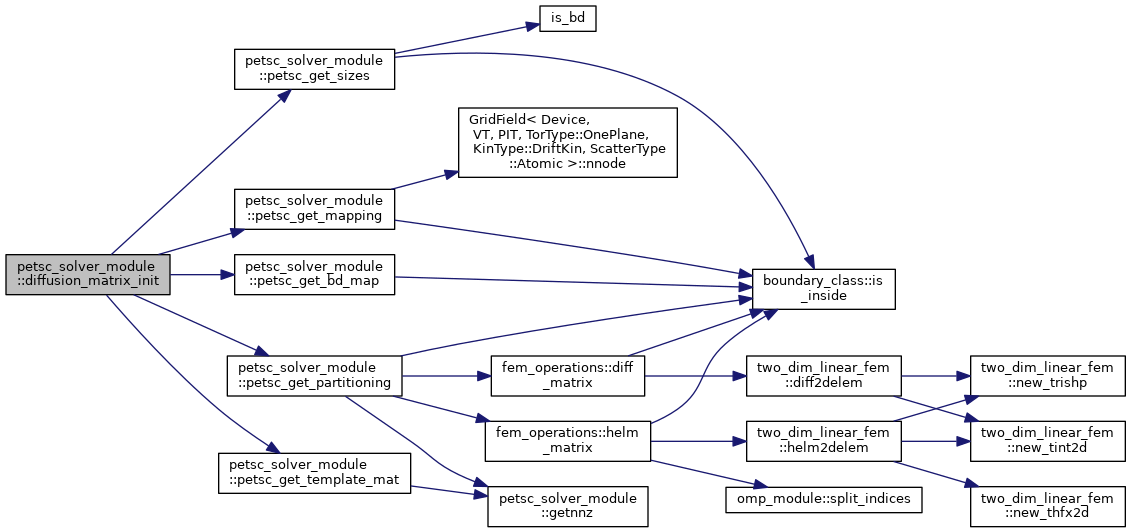
| subroutine | diffusion_matrix_init (diffusion_ts, grid, bc, solver_template_mat, ierr) |
| Set up a template matrix for XGC's anomalous diffusion time integrator. This is currently a system of either 4 (adiabatic electrons) or 7 (kinetic electrons) equations. Impurities are not supported yet. More... | |
| subroutine | set_adiabatic_blending_weights (grid, vec, xgc_petsc, ierr) |
| Sets values in a PETSc vector according to the blending function \(\alpha(\psi)\), where \(\alpha = 1\) in region 1 for \(\psi \le \psi_{\mathrm{in}}\), \(\alpha = \frac{\psi_{\textrm{out}} - \psi}{\psi_{\textrm{out}} - \psi_{\textrm{in}}}\) in region 1 for \(\psi_{\mathrm{in}} \lt \psi \lt \psi_{\mathrm{out}}\), and \(\alpha = 0\) in region 1 for \(\psi_{\mathrm{out}} \le \psi\) and outside of region 1. More... | |
| subroutine | create_helmholtz_solver (solver, solver_data, grid, bc, ierr) |
| Set up a KSP solver for a single Helmholtz-type equation (Poisson equation, Ampere's law) More... | |
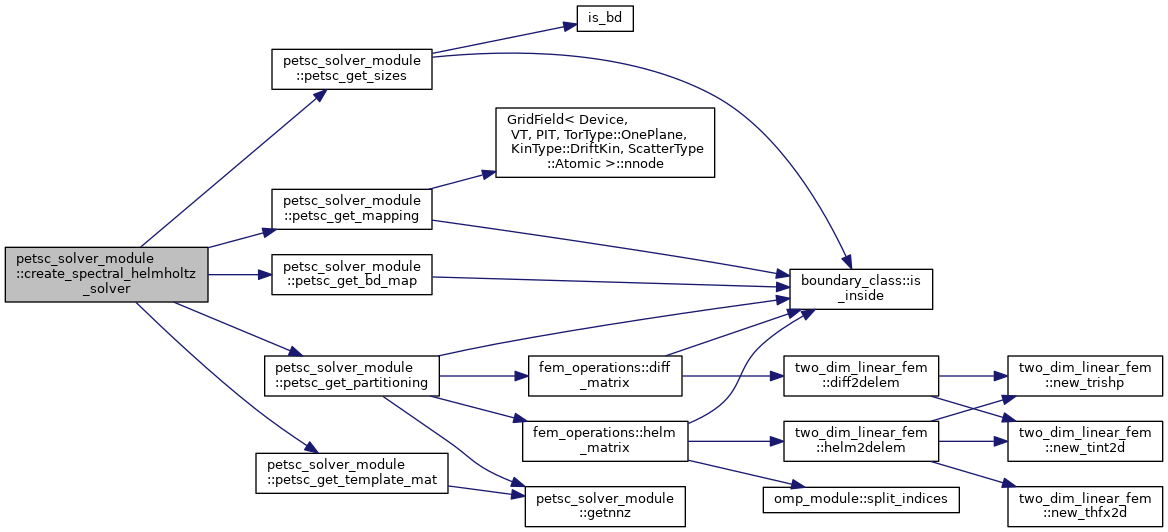
| subroutine | create_spectral_helmholtz_solver (solver, solver_data, grid, bc, ntor, ierr) |
| Set up a KSP solver for a single toroidal mode number component of Helmholtz-type equation (Poisson equation, Ampere's law) More... | |
| subroutine | init_helmholtz_solver (psn, grid, solver, solver_data, bd, comm, prefix, n_rhs_mat, is_axisym, is_spectral, is_ampere, is_ampere_cv, is_update) |
| Setup routine for the Poisson and the Ampere's law solvers This routine can be used for the initial set up, i.e., initializing the the distributed PETSc matrices and the KSP solver, and for updating exisiting solvers for the evolving background profiles. More... | |
| subroutine petsc_solver_module::create_1field_solver | ( | type(xgc_solver) | solver, |
| mat, | |||
| intent(out) | ierr | ||
| ) |
Creates a PETSc KSP solver for a one-block system, i.e., for a single equation on the XGC mesh.
| [in,out] | solver | XGC solver object, type(xgc_solver) |
| [in] | mat | Matrix from which to create KSP, Petsc Mat |
| [out] | ierr | PETSc Error code |

| subroutine petsc_solver_module::create_axisym_iter_solver | ( | type(xgc_solver) | solver, |
| intent(out) | ierr | ||
| ) |
Creates and sets up the PETSc KSP solver and preconditioner for inverting the axisymmetric Poisson operator.
| [in,out] | solver | XGC solver object, type(xgc_solver) |
| [out] | ierr | PETSc Error code |
| subroutine petsc_solver_module::create_helmholtz_solver | ( | type(xgc_solver) | solver, |
| type(solver_init_data) | solver_data, | ||
| type(grid_type), intent(in) | grid, | ||
| integer, dimension(grid%nnode), intent(in) | bc, | ||
| ierr | |||
| ) |
Set up a KSP solver for a single Helmholtz-type equation (Poisson equation, Ampere's law)
| [in,out] | solver | XGC solver object, type(xgc_solver), see module_psn.F90 |
| [in] | solver_data | Grid and magnetic field data needed for the perp. gradient |
| [in] | grid | XGC grid object, type(grid_type) |
| [in] | bc | Solver boundary mask, 1 if not in boundary vertex list |
| [out] | ierr | PETSc error code. |
| subroutine petsc_solver_module::create_spectral_helmholtz_solver | ( | type(xgc_solver) | solver, |
| type(solver_init_data) | solver_data, | ||
| type(grid_type), intent(in) | grid, | ||
| integer, dimension(grid%nnode), intent(in) | bc, | ||
| integer, intent(in) | ntor, | ||
| ierr | |||
| ) |
Set up a KSP solver for a single toroidal mode number component of Helmholtz-type equation (Poisson equation, Ampere's law)
| [in,out] | solver | XGC solver object, type(xgc_solver), see module_psn.F90 |
| [in] | solver_data | Grid and magnetic field data needed for the perp. gradient |
| [in] | grid | XGC grid object, type(grid_type) |
| [in] | bc | Solver boundary mask, 1 if not in boundary vertex list |
| [in] | ntor | Toroidal mode number for which to set up the solver |
| [out] | ierr | PETSc error code. |
| subroutine petsc_solver_module::diffusion_matrix_init | ( | type(xgc_ts) | diffusion_ts, |
| type(grid_type), intent(in) | grid, | ||
| integer, dimension(grid%nnode), intent(in) | bc, | ||
| intent(out) | solver_template_mat, | ||
| ierr | |||
| ) |
Set up a template matrix for XGC's anomalous diffusion time integrator. This is currently a system of either 4 (adiabatic electrons) or 7 (kinetic electrons) equations. Impurities are not supported yet.
| [in,out] | diffusion_ts | PETSc time integrator object (TS) for the diffusion solver |
| [in] | grid | XGC grid object, type(grid_type) |
| [in] | bc | Solver boundary mask, 1 if not in boundary vertex list |
| [out] | solver_template_mat | PETSc matrix with the template for the anomalous diffusion solver |
| [out] | ierr | PETSc error code. |

| subroutine petsc_solver_module::getnnz | ( | type(grid_type), intent(in) | grid, |
| nloc, | |||
| low, | |||
| high, | |||
| d_nnz, | |||
| o_nnz, | |||
| dimension(grid%nnode), intent(in) | xgc_petsc, | ||
| nglobal, | |||
| ierr | |||
| ) |
Computes the number of non-zero entries nnz per row in the rank-local rows.
| [in] | grid | XGC grid object, type(grid_type) |
| [in] | nloc | Number of equations on this rank, PetscInt |
| [in] | low | Global index of the first equation on this rank, PetscInt |
| [in] | high | Global index of the last equation on this rank, PetscInt |
| [out] | d_nnz | Number of non-zero entries in "diagonal" local part of the matrix |
| [out] | o_nnz | Number of non-zero entries in the "off-diagnonal" local part of the matrix. |
| [in] | xgc_petsc | Mapping between XGC grid vertices and PETSc equations |
| [in] | nglobal | Total number of equations, PetscInt |
| [in] | ierr | PETSc error code |
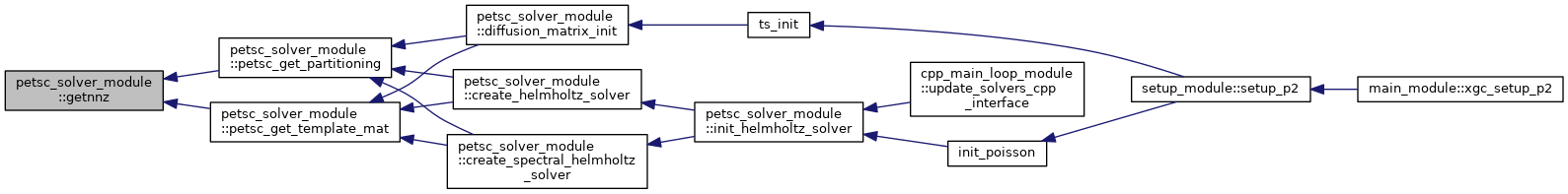
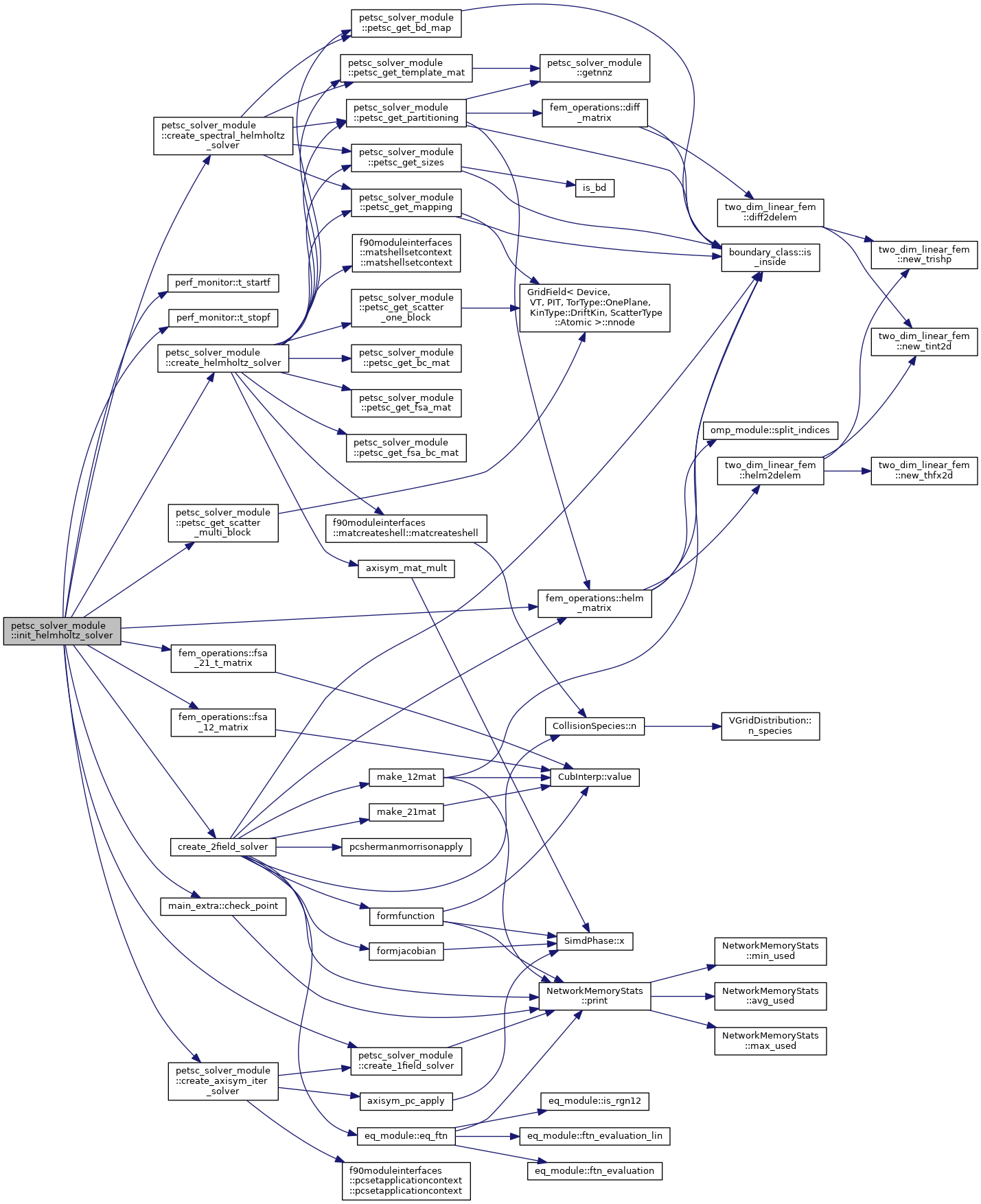
| subroutine petsc_solver_module::init_helmholtz_solver | ( | type(psn_type), intent(inout) | psn, |
| type(grid_type), intent(in) | grid, | ||
| type(xgc_solver), intent(inout) | solver, | ||
| type(solver_init_data), intent(in) | solver_data, | ||
| integer, dimension(grid%nnode), intent(in) | bd, | ||
| integer, intent(in) | comm, | ||
| character(*), intent(in) | prefix, | ||
| integer, intent(in) | n_rhs_mat, | ||
| logical, intent(in) | is_axisym, | ||
| logical, intent(in) | is_spectral, | ||
| logical, intent(in) | is_ampere, | ||
| logical, intent(in) | is_ampere_cv, | ||
| logical, intent(in) | is_update | ||
| ) |
Setup routine for the Poisson and the Ampere's law solvers This routine can be used for the initial set up, i.e., initializing the the distributed PETSc matrices and the KSP solver, and for updating exisiting solvers for the evolving background profiles.
| [in,out] | psn | XGC field object, type(psn_type) |
| [in] | grid | XGC grid object, type(grid_type) |
| [in,out] | solver | XGC solver object, type(xgc_solver_type) |
| [in] | solver_data | Data required for solver setup (n, T, etc.), type(solver_init_data) |
| [in] | bd | Solver boundary mask, 1 if not in boundary vertex list |
| [in] | comm | Communicator of the solver, integer |
| [in] | prefix | Prefix string for the solver, character |
| [in] | n_rhs_mat | Number of right-hand side operators (1 or 2), integer |
| [in] | is_axisym | Set up for axisymmetric solver if .true., logical |
| [in] | is_spectral | Set up for spectral solver if .true., logical |
| [in] | is_ampere | Set up Ampere's law solver if .true., logical |
| [in] | is_ampere_cv | Set up control-variate Ampere's law solver, logical if .true., logical |
| [in] | is_update | If .true. update an already initialized solver, logical |
| subroutine petsc_solver_module::init_ksp_comm | ( | integer, intent(in) | nnode | ) |
Initializes an MPI communicator for use with PETSc KSP solves (Poisson, Ampere). The size of the KSP comm group is set such that, if possible, the number of equations per MPI rank is larger than 5000, which is roughly the weak scaling rollover.
| [in] | nnode | Number of mesh vertices per poloidal plane, integer |
| subroutine petsc_solver_module::petsc_get_bc_mat | ( | type(xgc_solver) | solver, |
| intent(in) | n_eq_tot, | ||
| intent(in) | n_eq_loc, | ||
| intent(out) | ierr | ||
| ) |
Creates the (empty) LHS and RHS matrices for Dirichlet boundary conditions.
| [in,out] | solver | XGC solver object |
| [in] | n_eq_tot | Number of equations to solve, PetscInt |
| [in] | n_eq_loc | Number of equations on this rank, PetscInt |
| [out] | ierr | PETSc error code |

| subroutine petsc_solver_module::petsc_get_bd_map | ( | type(grid_type), intent(in) | grid, |
| integer, dimension(grid%nnode), intent(in) | bc, | ||
| intent(in) | n_boundary, | ||
| dimension(grid%nnode), intent(in) | petsc_xgc_bd, | ||
| dimension(n_boundary), intent(out) | petsc_bd_xgc | ||
| ) |
Generate mapping from PETSc boundary conditions to XGC vertices. This requires the results of petsc_get_sizes because the number of boundary conditions is not known a priori.
| [in] | grid | XGC solver mesh, type(grid_type) |
| [in] | bc | Solver boundary mask, 1 if not in boundary vertex list |
| [in] | n_boundary | Number of boundary values, PetscInt |
| [in] | petsc_xgc_bd | Mapping from XGC vertices to PETSc boundary conditions |
| [out] | petsc_bd_xgc | Mapping fron PETSc boundary conditions to XGC vertices |
| subroutine petsc_solver_module::petsc_get_fsa_bc_mat | ( | type(xgc_solver) | solver, |
| intent(out) | ierr | ||
| ) |
creates and allocates boundary/surface FSA matrices
| [in,out] | solver | XGC solver object |
| [out] | ierr | PETSc error code |

| subroutine petsc_solver_module::petsc_get_fsa_mat | ( | type(xgc_solver) | solver, |
| intent(in) | n_eq_tot, | ||
| intent(in) | n_eq_loc, | ||
| intent(out) | ierr | ||
| ) |
creates and allocates interior/surface FSA matrices
| [in,out] | solver | XGC solver object |
| [in] | n_eq_tot | Number of equations to solve, PetscInt |
| [in] | n_eq_loc | Number of equations on this rank, PetscInt |
| [out] | ierr | PETSc error code |

| subroutine petsc_solver_module::petsc_get_mapping | ( | integer, intent(in) | nnode, |
| integer, dimension(nnode), intent(in) | bc, | ||
| integer, intent(in) | comm, | ||
| integer, intent(in) | num_pe, | ||
| intent(in) | n_eq_tot, | ||
| intent(in) | n_eq_loc, | ||
| dimension(n_eq_tot), intent(in) | xgc_proc, | ||
| dimension(0:num_pe), intent(inout) | proc_eq, | ||
| dimension(nnode), intent(out) | xgc_petsc, | ||
| dimension(n_eq_tot), intent(out) | petsc_xgc, | ||
| dimension(n_eq_loc), intent(out) | petscloc_xgc | ||
| ) |
Based on the Parmetis partition of the XGC domain, updates the mappings between XGC vertices and PETSc equations.
| [in] | nnode | Number of XGC vertices, integer |
| [in] | bc | Solver boundary mask, 1 if not in boundary vertex list |
| [in] | comm | MPI comunicator for distributed matrix solver, MPI_comm |
| [in] | n_eq_tot | Total number of PETSc equations, PetscInt |
| [in] | n_eq_loc | Local number of equations on the current rank, PetscInt |
| [in] | xgc_proc | Mapping from equation index to XGC rank, PetscInt |
| [in] | proc_eq | Mapping from XGC process to PETSc equation, integer |
| [out] | xgc_petsc | Mapping from XGC vertices to PETSc equation, integer |
| [out] | petsc_xgc | Mapping from PETSc equation index to XGC vertices, integer |
| [out] | petscloc_xgc | Mapping from local PETSc eq. to XGC vertices, integer |
| subroutine petsc_solver_module::petsc_get_partitioning | ( | type(grid_type), intent(in) | grid, |
| integer, dimension(grid%nnode), intent(in) | bc, | ||
| type(solver_init_data), intent(in) | solver_data, | ||
| integer, intent(in) | comm, | ||
| integer, intent(in) | num_pe, | ||
| intent(in) | n_eq_tot_in, | ||
| logical, intent(in) | set_diffusion_matrix, | ||
| dimension(grid%nnode), intent(in) | xgc_petsc, | ||
| dimension(grid%nnode), intent(in) | petsc_xgc_bd, | ||
| intent(out) | n_eq_loc, | ||
| dimension(n_eq_tot_in), intent(out) | xgc_proc_out, | ||
| dimension(0:num_pe), intent(out) | proc_eq, | ||
| intent(out) | ierr | ||
| ) |
Generates a domain partitioning of part of the XGC mesh used by a solver (as defined by the boundary conditions bc) using Parmetis and generates a mapping between PETSc equation index and XGC MPI ranks (those in the communicator comm used by the solver).
| [in] | grid | XGC solver mesh, type(grid_type) |
| [in] | bc | Solver boundary mask, 1 if not in boundary vertex list |
| [in] | comm | MPI communicator used by the solver, integer |
| [in] | num_pe | Number of MPI ranks in communicator comm, integer |
| [in] | n_eq_tot_in | Total number of equations (i.e. number of vertices in the solver), PetscInt |
| [in] | set_diffusion_matrix | Whether to set diffusion or Poisson/Ampere matrix |
| [out] | n_eq_loc | Number of equations on the current rank, PetscInt |
| [out] | xgc_proc | Mapping from PETSc equation to XGC rank, PetscInt |
| [out] | proc_eq | Mapping from XGC_process to PETSc equation number (in form of the boundaries of the domain partitioning) |
| [out] | ierr | Error code |
| subroutine petsc_solver_module::petsc_get_scatter_multi_block | ( | integer, intent(in) | nnode, |
| intent(in) | ksp, | ||
| mat_all_blocks, | |||
| mat_one_block, | |||
| intent(in) | blocksize, | ||
| character(*), dimension(0:blocksize-1), intent(in) | varnames, | ||
| intent(in) | n_eq_tot, | ||
| intent(in) | n_eq_loc, | ||
| dimension(n_eq_loc), intent(in) | petscloc_xgc, | ||
| dimension(n_eq_tot), intent(in) | petsc_xgc, | ||
| iss, | |||
| to_petsc, | |||
| from_petsc, | |||
| intent(out) | ierr | ||
| ) |
Sets up a PETSc scatter object for a (2D) multi-block field-split solver.
| [in] | nnode | Number of XGC mesh vertices |
| [in] | ksp | PETSc Krylov sub-space solver object, KSP |
| [in] | mat_all_blocks | Global matrix with all solver blocks, Mat |
| [in] | mat_one_block | Matrix of one single block, Mat |
| [in] | blocksize | Number of blocks, PetscInt |
| [in] | varnames | Names of the variables represented by each block, char |
| [in] | n_eq_tot | Number of equations in one block, PetscInt |
| [in] | n_eq_loc | Number of local equations in one block, PetscInt |
| [in] | petscloc_xgc | Rank-local mapping from PETSc equations to XGC vertices, PetscInt |
| [in] | petsc_xgc | Global mapping from PETSc equations to XGC vertices, PetscInt |
| [out] | iss | Index sets for mapping single blocks into the global matrix, IS |
| [out] | from_petsc | VecScatter object for scattering PETSc results to an XGC field, VecScatter |
| [out] | to_petsc | VecScatter object for scattering XGC vectors into the global PETSc block matrix |
| [out] | ierr | PETSc error code, PetscErrorCode |
| subroutine petsc_solver_module::petsc_get_scatter_one_block | ( | type(xgc_solver) | solver, |
| integer, intent(in) | nnode, | ||
| intent(in) | n_eq_tot, | ||
| intent(in) | n_eq_loc, | ||
| dimension(n_eq_tot), intent(in) | petsc_xgc, | ||
| intent(out) | ierr | ||
| ) |
Generate PETSc scatter mapping from XGC vertices to PETSc equation for a simple solver with one equation (block). Also sets up the corresponding LHS and RHS PETSc vectors.
| [in,out] | solver | XGC solver object |
| [in] | nnode | Number of XGC mesh vertices |
| [in] | n_eq_tot | Total number of PETSc equations, PetscInt |
| [in] | n_eq_loc | Number of PETSc equations on this rank |
| [in] | petsc_xgc | Mapping from PETSc equation index to XGC mesh vertices |
| [out] | ierr | PETSc error code |

| subroutine petsc_solver_module::petsc_get_sizes | ( | type(grid_type), intent(in) | grid, |
| integer, dimension(grid%nnode), intent(in) | bc, | ||
| intent(out) | n_equation, | ||
| intent(out) | n_boundary, | ||
| dimension(grid%nnode), intent(out) | xgc_petsc, | ||
| petsc_xgc_bd | |||
| ) |
Calculate (i) the number of equations (vertices) of the solver, (ii) the number of XGC boundary vertices included in the solver, (iii) (preliminary) mapping from XGC vertices to PETSc equations, (iv) mapping from XGC vertices to PETSc boundary conditions, all based the XGC boundary object passed as input.
| [in] | grid | XGC solver mesh, type(grid_type) |
| [in] | bc | Solver boundary mask, 1 if not in boundary vertex list |
| [out] | n_equation | Number of PETSc equations (matrix rows), integer |
| [out] | n_boundary | Number of boundary conditions/vertices, integer |
| [out] | xgc_petsc | (Preliminary) mapping from XGC vertices to PETSc equations |
| [out] | petsc_xgc_bd | Mapping from XGC vertices to PETSc boundary conditions |
| subroutine petsc_solver_module::petsc_get_template_mat | ( | type(grid_type), intent(in) | grid, |
| integer, intent(in) | comm, | ||
| intent(in) | n_eq_tot, | ||
| intent(in) | n_eq_loc, | ||
| dimension(grid%nnode), intent(in) | xgc_petsc, | ||
| intent(out) | solver_template_mat, | ||
| ierr | |||
| ) |
Uses pre-computed (petsc_get_partitioning) local matrix sizes and XGC vertex to PETSc equation mapping to set up a blank template matrix with sufficient pre-allocated memory.
| [in] | grid | XGC grid object, type(grid_type) |
| [in] | comm | MPI communicator for template matrix, MPI_comm |
| [in] | n_eq_tot | Total number of equations, PetscInt |
| [in] | n_eq_loc | Number of local equations on the current rank, PetscInt |
| [in] | xgc_petsc | Mapping from XGC vertices to PETSc equation index |
| [out] | solver_template_mat | Template matrix with the desired partitioning and sufficient preallocated memory. |
| [out] | ierr | Error code |
| subroutine petsc_solver_module::petsc_to_xgc | ( | intent(in) | nnode, |
| intent(in) | blocksize, | ||
| dimension(0:blocksize-1), intent(in) | from_petsc, | ||
| intent(in) | field_petsc, | ||
| real (kind=8), dimension(nnode,blocksize), intent(inout) | field_xgc, | ||
| intent(out) | ierr | ||
| ) |
This routine scatters values from the distributed vector compatible with the block-matrix of the 2D diffusion model to local variables on the XGC solver grid.
| [in] | nnode | Number of vertices per plane in XGC mesh, PetscInt |
| [in] | blocksize | Number of equations, PetscInt |
| [in] | from_petsc | VecScatter object for moving data from PETSc Vec to XGC mesh data, VecScatter |
| [in] | field_petsc | PETSc vector with the data to be scattered, PETSc Vec |
| [in,out] | field_xgc | XGC mesh array to which data is scattered, real8 |
| [out] | ierr | Error code, PetscErrorCode |
| subroutine petsc_solver_module::petsc_to_xgc_bd | ( | integer | nn, |
| type(xgc_solver) | this, | ||
| x, | |||
| xvec | |||
| ) |
Scatters a PETSc vector to XGC boundary data.
| [in] | nn | size of x, integer |
| [in] | this | XGC solver data object, type(xgc_solver) |
| [out] | x | XGC scalar field (defined on boundary vertices) |
| [in] | xvec | Petsc vector, Vec |
| subroutine petsc_solver_module::set_adiabatic_blending_weights | ( | type(grid_type), intent(in) | grid, |
| vec, | |||
| dimension(grid%nnode), intent(in) | xgc_petsc, | ||
| intent(out) | ierr | ||
| ) |
Sets values in a PETSc vector according to the blending function \(\alpha(\psi)\), where \(\alpha = 1\) in region 1 for \(\psi \le \psi_{\mathrm{in}}\), \(\alpha = \frac{\psi_{\textrm{out}} - \psi}{\psi_{\textrm{out}} - \psi_{\textrm{in}}}\) in region 1 for \(\psi_{\mathrm{in}} \lt \psi \lt \psi_{\mathrm{out}}\), and \(\alpha = 0\) in region 1 for \(\psi_{\mathrm{out}} \le \psi\) and outside of region 1.
| [in] | grid | XGC grid object, type(grid_type) |
| [in,out] | vec | PETSc vector to store blending weights, Vec |
| [in] | xgc_petsc | Mapping from XGC vertex index to PETSc equation number, PetscInt |
| [out] | ierr | error code, PetscErrorCode |
| subroutine petsc_solver_module::xgc_to_petsc | ( | intent(in) | nnode, |
| intent(in) | blocksize, | ||
| dimension(0:blocksize-1), intent(in) | to_petsc, | ||
| field_petsc, | |||
| real (kind=8), dimension(nnode,blocksize), intent(in) | field_xgc, | ||
| intent(out) | ierr | ||
| ) |
This routine scatters values from local variable on the XGC solver grid to the distributed vector compatible with the block-matrix of the 2D diffusion model.
| [in] | nnode | Number of vertices per plane in XGC mesh, PetscInt |
| [in] | blocksize | Number of equations, PetscInt |
| [in] | from_petsc | VecScatter object for moving data from PETSc Vec to XGC mesh data, VecScatter |
| [in] | field_petsc | PETSc vector with the data to be scattered, PETSc Vec |
| [in,out] | field_xgc | XGC mesh array to which data is scattered, real8 |
| [out] | ierr | Error code, PetscErrorCode |
| subroutine petsc_solver_module::xgc_to_petsc_bd | ( | integer | nn, |
| type(xgc_solver) | this, | ||
| x, | |||
| xvec | |||
| ) |
Scatters XGC boundary data to a PETSc vector.
| [in] | nn | size of x, integer |
| [in] | this | XGC solver data object, type(xgc_solver) |
| [out] | x | XGC scalar field (defined on boundary vertices) |
| [in] | xvec | Petsc vector, Vec |
 1.8.5
1.8.5